Post-Translational Modification regulates DNA repair and suggests new therapeutic targets.
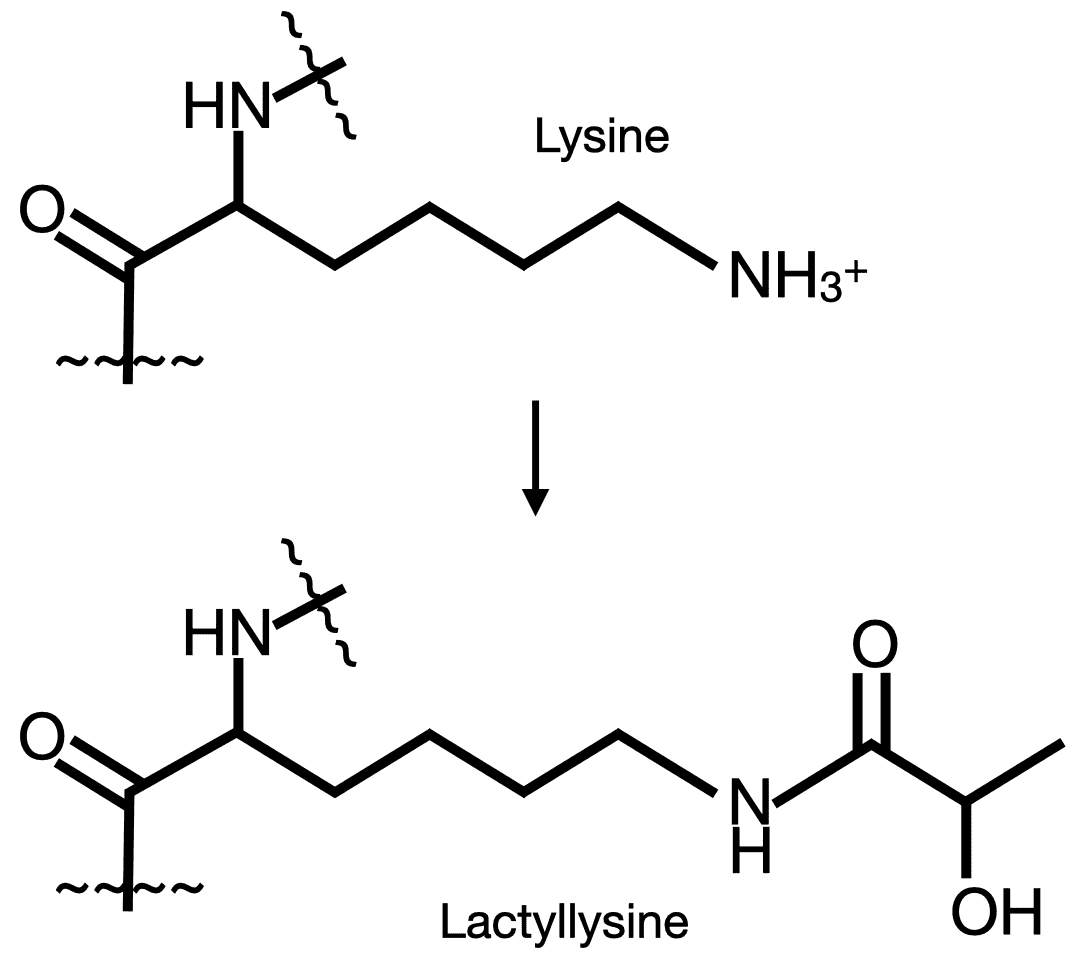
A host of protein post-translational modifications (PTMs) have been discovered and play important roles in cellular regulation and metabolism. Recently, a new PTM was discovered that modifies protein lysine to lactyllysine in histones and plays an important role in regulation of gene expression (Zhang, D., Tang, Z., Huang, H., Zhou, G., Cui, C., Weng, Y., et al. (2019). Metabolic regulation of gene expression by histone lactylation. Nature 574 (7779), 575–580. https://doi.org/10.1038/s41586-019-1678-1). Since this original finding, lactylation has been observed in many proteins across domains of life and suggests important cellular roles.
This week in Cell, Chen et al. report a new function of lactylation in regulation of DNA repair. They show that specific lactylation of the DNA repair MRE11 by CBP acetyltransferase promotes DNA binding and facilitates end resection during homologous recombination. They demonstrated that blocking MRE11 lactylation inhibits homologous recombination and sensitized cancer cells to cisplatin and PARPi suggesting new therapeutic targets.
Read the paper here:
Chen et al. “Metabolic regulation of homologous recombination repair by MRE11 lactylation.” (2023) Cell. Dec 18:S00-3-8674(23)01276-X https://doi.org/10.1016/j.cell.2023.11.022
